
Jack Foulkes
Trainee Clinical Scientist in Imaging with Non-Ionising Radiation, St George's University Hospitals NHS Foundation Trust

Jack Foulkes 4 week STP elective
15 May 2023
I’m a trainee clinical scientist in medical physics, specializing in MRI, currently in my 3rd and final year of study on the NHS Scientist Training Programme (STP). I was slightly nervous to apply to the CSC for my elective placement, as I had very limited experience in coding so far during my career, and no experience whatsoever in professional software development! However, my experience going in was incredibly warm and welcoming. I’ve felt very privileged so far to spend a month with the CSC team at GSTT, during which I was able to spend some time with each member of the team and grill them about the projects they’re involved in. My aims going in were to get more experience in proper software development and testing methodologies, and also to learn more about the new applications of artificial intelligence (AI) and machine learning (ML) within an increasingly digitized healthcare system.
In my first week I was able to attend one of the bi-weekly 10X workshops, in which the CSC team all meet together in person to discuss an issue relevant to their work. This week workshop was focused on an issue brought to the team by the general manager for the radiology department at an NHS hospital, who was struggling to find the cause of a huge backlog in radiology reporting. After a presentation by the general manager, instead of jumping to offer solutions the team lead (Haris Shuaib) encouraged the other members to more clearly define the exact parameters of the problem. Haris engaged the team in a system mapping exercise, during which the whole system that you’re trying to understand is mapped out in a process diagram, allowing the potential flaws in the various processes to be identified more easily.
An online Miro board was used to create a process diagram, such that those team members joining online could engage in the activity, and all team members could lay out their suggested ideas and questions about the processes involved. The aim of this exercise was to establish the simplest, most cost-effective solution to the problem, before suggesting the application of AI. Once a set of solutions had been generated, the solutions were all placed inside an importance/difficulty matrix (see Figure 2) to help decide on which solution to focus on in future.
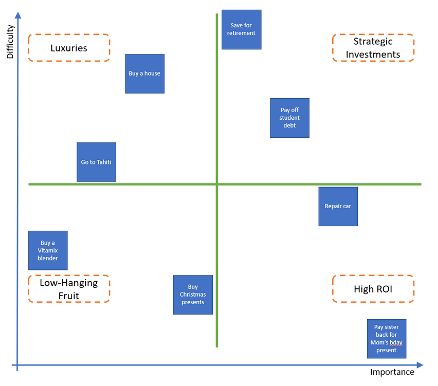
I was impressed that the team was able to prioritise cost-effectiveness for the NHS over a more technologically-driven approach which I had perhaps expected. It was really exciting to see huge problems in the healthcare sector being tackled head on by such a small group of people; this experience gave me much more confidence in the efficacy of scientists in the NHS more broadly.
During my placement I was able to spend some time with Dika Vilic, who walked me through the capabilities of the XNAT informatics platform, which is used at GSTT for retrieval and anonymisation of medical images for research and ML applications. One of the many advantages of XNAT is that the code is open source and extensible, meaning that one can effectively customize the platform to the individual needs of the department. With Dika’s help, I was able to form a step-by-step plan for how to get an installation of XNAT working within an NHS radiology IT network, which will be really valuable to facilitate research in my next role when I finish the STP.
I spoke with Tom Roberts about the new application TotalSegmentator, which uses an ML trained algorithm to automatically segment organs in CT or MR images used for radiotherapy treatment planning. This application will be available on the open source AI platform referred to as AIDE. Tom is hoping to validate the application prior to release, by comparing the output to segmentation data created by other clinicians. This conversation helped me to understand the importance of training these algorithms on data derived from patients with a variety of racial and ethnic backgrounds, to help ensure that the application will have a good performance across a diverse range of patients, more reflective of the society we live in. It also gave me some insight into the amount of work needed to preprocess the data that is used for training to ensure that it is usable, which is a separate process to the curation of data.
I also spent some time writing unit tests for the Hazen project, which is used for quality assurance analysis in medical imaging. Initially I found it very difficult to work within a professional integrated development environment (PyCharm), and it took me a while to get my head around GitHub, but the team were really supportive as soon as I asked for help during one of the twice-weekly stand-up meetings. I had a really useful interview with Haris Shuaib, who talked to me about the practice of test-driven development (i.e. writing tests before the code!) which has since revolutionized my understanding of writing software.
I’m now in my last week here at CSC, and my predominant feeling is that I wish I had more time here. I’ve been overwhelmed with the level of innovation going on within this team, and I’ve learned so much about the benefits and difficulties of applying artificial intelligence to solve problems in healthcare, as well as a bunch of great tools for professional software development. Although the learning curb has been sharp, my main takeaway is that anyone is capable of learning to develop the skills necessary to develop good quality software to solve problems in the NHS.