XNAT

XNAT is an open-source imaging informatics platform for storing and managing medical images and associated data for research purposes. At GSTT, we use XNAT to retrieve, anonymise, and manage medical imaging and non-imaging data for purposes of AI development, AI evaluation, and for data provision for various research projects which are run at GSTT or in collaboration with GSTT.
XNAT at GSTT is currently connected to 3 PACS systems (radiology, foetal cardiology and radiotherapy PACS). Data anonymisation processes include DICOM meta-data removal, face blurring and removal of burnt-in data. Data used for AI evaluation are pushed to relevant DICOM nodes using a Python script; data used for AI development are fetched by MLOPs via XNAT’s REST API, and data are sent to external collaborators via pathways agreed in ethics approval of each individual project.
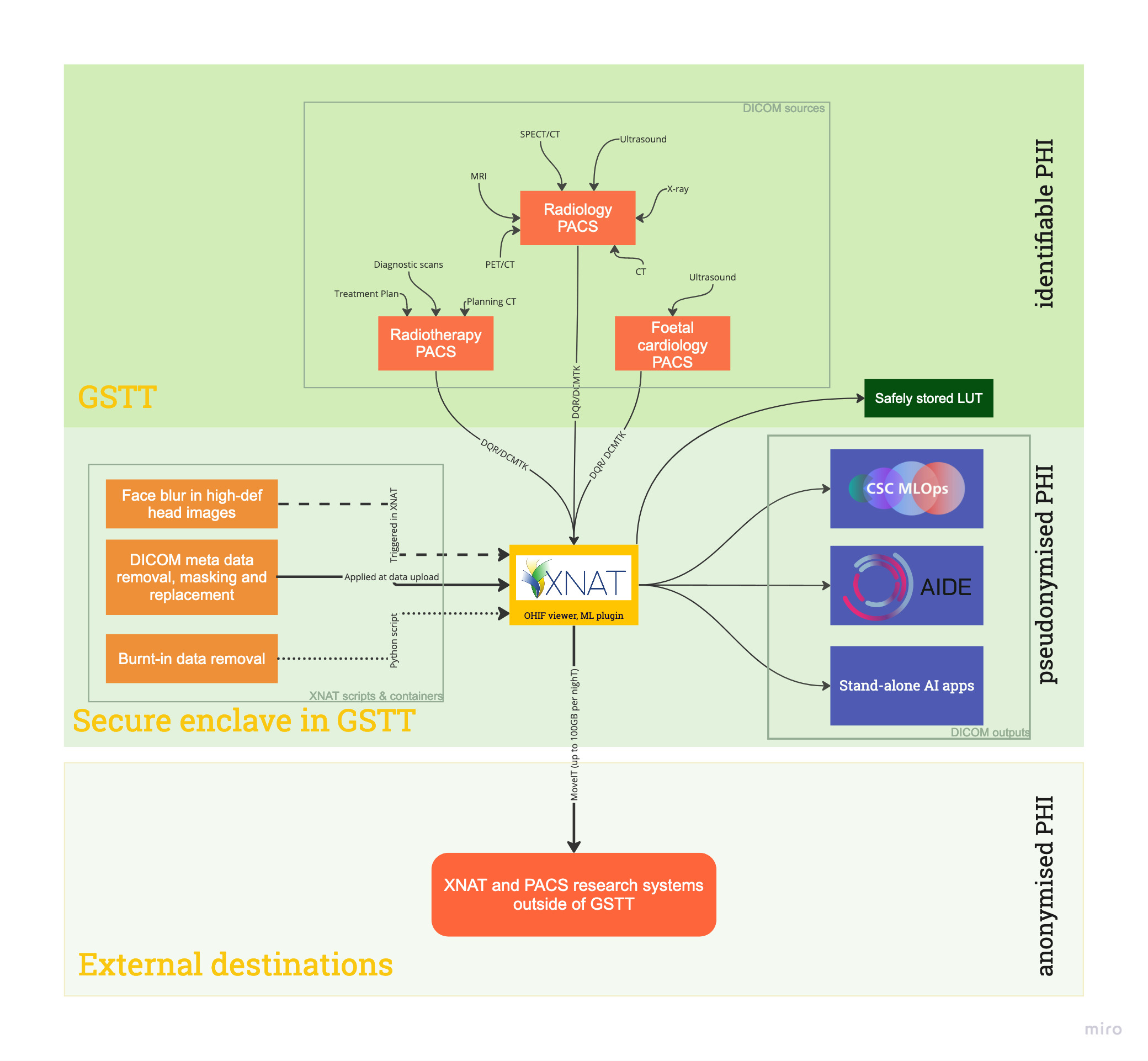
Using XNAT for data anonymisation and management allows fast and automated retrieval of research data, allows for good quality control and application of highest standards of anonymisation to protect patient privacy. Data visibility is granted explicitly, so only collaborators on a certain project have data visibility. Lastly, access to XNAT is strictly controlled and regularly reviewed.
XNAT is only available to GSTT employees with access to GSTT network and before access can be granted an online teaching and training session is held. If you need access to XNAT, please contact the CSC team.
For a wealth of information, including our SOPs, anonymisation scripts, data retrieval and fetching scripts and similar, please visit our XNAT GitHub repository.
If you would like to us to help you retrieve data for your project and perform the relevant anonymisation process, please get in touch!